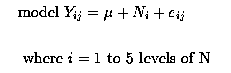
Many factors are quantitative in nature and cause us to incorporate them in a model as regression effects. For example, the data presented earlier relating leaf burn in tobacco plants to the Nitrogen, Chloride and Phosphorus content. Nitrogen%, Chloride% and Phosphorus% were considered as the independent variables to explain variation in leaf burn. They were considered as Quantitative/Covariate Regression variables. We were looking at the effect of each unit increase (or decrease) in N% on leaf burn; and similarly for the other variables. Nitrogen%, Cl% and P% are quantitative; 5% is half of 10%, which is half of 20%, etc.
Many factors are qualitative in nature and cause us to incorporate them in a model as classification effects (qualitative variables with classification 'levels'). Classification, or qualitative, variables cannot be considered as quantities. For example, if we are looking at the growth rate of male and female pigs, we can call them M and F, or 1 and 2 respectively (or 2 and 1 also). The 'levels' of sex (M and F) are qualitative in nature, there is no 'quantity' of sex associated with being a Male or a Female. The labels M and F are 'qualities' and serve only to identify or differentiate which animals are of one sex or the other.
The SAS analyses of the data presented in the class notes, with 5 diets and 5 rats per Diet is shown here, Rats!.
Model fitting diet as 5 classes (4 degrees of freedom), a regular classification model, CRD.
| The GLM Procedure |
| Class Level Information | ||
| Class | Levels | Values |
| diet | 5 | 1 2 3 4 5 |
| Number of observations | 25 |
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
| Model | 4 | 1985.674400 | 496.418600 | 148.53 | <.0001 |
| Error | 20 | 66.844000 | 3.342200 | ||
| Corrected Total | 24 | 2052.518400 |
We can see here the ANOVA table produced from the data and SAS PROC GLM code given in the course notes. Note the effect of Diet is statistically significant, F-ratio = 148.53. This is the effect of Diets (5 'levels', 4 degrees of freedom) over and above the Mean, R(Diet|µ)
| R-Square | Coeff Var | Root MSE | wt Mean |
| 0.967433 | 4.479927 | 1.828168 | 40.80800 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
| diet | 4 | 1985.674400 | 496.418600 | 148.53 | <.0001 |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
| diet | 4 | 1985.674400 | 496.418600 | 148.53 | <.0001 |
| Parameter | Estimate | Standard Error | t Value | Pr > |t| |
| mu+d1 | 23.9600000 | 0.81758180 | 29.31 | <.0001 |
| mu+d2 | 39.5800000 | 0.81758180 | 48.41 | <.0001 |
| mu+d3 | 45.8400000 | 0.81758180 | 56.07 | <.0001 |
| mu+d4 | 48.1600000 | 0.81758180 | 58.91 | <.0001 |
| mu+d5 | 46.5000000 | 0.81758180 | 56.88 | <.0001 |
| The GLM Procedure |
| Least Squares Means |
| Adjustment for Multiple Comparisons: Scheffe |
| diet | wt LSMEAN | Standard Error | Pr > |t| | LSMEAN Number |
| 1 | 23.9600000 | 0.8175818 | <.0001 | 1 |
| 2 | 39.5800000 | 0.8175818 | <.0001 | 2 |
| 3 | 45.8400000 | 0.8175818 | <.0001 | 3 |
| 4 | 48.1600000 | 0.8175818 | <.0001 | 4 |
| 5 | 46.5000000 | 0.8175818 | <.0001 | 5 |
|
Least Squares Means for effect diet Pr > |t| for H0: LSMean(i)=LSMean(j) Dependent Variable: wt |
|||||
| i/j | 1 | 2 | 3 | 4 | 5 |
| 1 | <.0001 | <.0001 | <.0001 | <.0001 | |
| 2 | <.0001 | 0.0008 | <.0001 | 0.0003 | |
| 3 | <.0001 | 0.0008 | 0.4275 | 0.9872 | |
| 4 | <.0001 | <.0001 | 0.4275 | 0.7253 | |
| 5 | <.0001 | 0.0003 | 0.9872 | 0.7253 | |
Model fitting a 4th order polynomial (linear, quadratic, cubic and quartic regressions), providing EXACTLy the same fit, SSRm, etc.
| The GLM Procedure |
| Number of observations | 25 |
| Dependent Variable: wt |
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
| Model | 4 | 1985.674400 | 496.418600 | 148.53 | <.0001 |
| Error | 20 | 66.844000 | 3.342200 | ||
| Corrected Total | 24 | 2052.518400 |
| R-Square | Coeff Var | Root MSE | wt Mean |
| 0.967433 | 4.479927 | 1.828168 | 40.80800 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
| diet | 1 | 1439.697800 | 1439.697800 | 430.76 | <.0001 |
| diet*diet | 1 | 529.375000 | 529.375000 | 158.39 | <.0001 |
| diet*diet*diet | 1 | 14.472200 | 14.472200 | 4.33 | 0.0505 |
| diet*diet*diet*diet | 1 | 2.129400 | 2.129400 | 0.64 | 0.4341 |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
| diet | 1 | 15.33436985 | 15.33436985 | 4.59 | 0.0447 |
| diet*diet | 1 | 5.36020200 | 5.36020200 | 1.60 | 0.2199 |
| diet*diet*diet | 1 | 2.87482134 | 2.87482134 | 0.86 | 0.3648 |
| diet*diet*diet*diet | 1 | 2.12940000 | 2.12940000 | 0.64 | 0.4341 |
| Parameter | Estimate | Standard Error | t Value | Pr > |t| |
| Intercept | -11.90000000 | 12.95293171 | -0.92 | 0.3692 |
| diet | 50.97166667 | 23.79644517 | 2.14 | 0.0447 |
| diet*diet | -18.06250000 | 14.26275753 | -1.27 | 0.2199 |
| diet*diet*diet | 3.17833333 | 3.42696943 | 0.93 | 0.3648 |
| diet*diet*diet*diet | -0.22750000 | 0.28501584 | -0.80 | 0.4341 |
Here we see the data analysed using a 4th order polynomial, diet, diet2, diet3 and diet4, and note the we obtain EXACTLY the same F-ratio for the effects. This is because R(diet ,diet2, diet3 and diet4 |µ) is an equivalence for the model with diets as 5 classes.
Molde fitting BOTH regressions (linear and quadratic) and classification, with the objective of seeing the marginal improvement by the classes over and above the regressions. An 'Over-paramereized Model'.
| The GLM Procedure |
| Class Level Information | ||
| Class | Levels | Values |
| diet | 5 | 1 2 3 4 5 |
| Number of observations | 25 |
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
| Model | 4 | 1985.674400 | 496.418600 | 148.53 | <.0001 |
| Error | 20 | 66.844000 | 3.342200 | ||
| Corrected Total | 24 | 2052.518400 |
| R-Square | Coeff Var | Root MSE | wt Mean |
| 0.967433 | 4.479927 | 1.828168 | 40.80800 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
| d | 1 | 1439.697800 | 1439.697800 | 430.76 | <.0001 |
| d*d | 1 | 529.375000 | 529.375000 | 158.39 | <.0001 |
| diet | 2 | 16.601600 | 8.300800 | 2.48 | 0.1088 |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
| d | 0 | 0.00000000 | . | . | . |
| d*d | 0 | 0.00000000 | . | . | . |
| diet | 2 | 16.60160000 | 8.30080000 | 2.48 | 0.1088 |
Here we see another model fitted, this one may look a little strange, but in fact it is a combination of the regression and classification. It allows us to test whether the classification provides a statistically significant improvement in the fit of the model ovef and above the linear and quadratic regressions. What we have done (check the PROC GLM statements, is to include the effect d and d2, which correspond to linear and quadratic effects of diet since they were not declared as CLASS variables, BEFORE Diet, which was declared as a class variable. This means that when we come to compute the marginal effect of Diet over and above the other effects in the model (the advantage of the Type III Sums of Squares), we are looking for R(Diet|µ, d, d2). But, and note that this is the 'tricky' bit, Diet is equal to d, d2, d3 and d4, the 4th order polynomial. Thus this is effectively equivalent to R(d, d2, d3, d4 | µ,d, d2). Now we have d and d2 being fitted twice! Thus when we come to fit Diet, as 5 classes/4 degrees of freedom, we have already fitted d and d2, and hence there is nothing new to fit for them. Therefore GLM realises that this is happening (because the rank is 2 d.f. less than one would otherwise think) and only provides us with 2 extra d.f. for Diet, othe and above what we have already effectively fitted; together with the Marginal Sums of Squares. Thus we get a direct and immediate test of whether the 5 classes was better than the linear and quadratic regressions. In this case we can see the the F-ratio for Diet over and above the linear and quadratic regressions was 2.48, which is NOT Statistically Significant. Thus the 5 classes DOES NOT provide a better fit, and hence we can simplify our model to be:
Consider the case when we have data where the independent variable (X) can be considered as a quantitative variable and /or as a classification variable with 'levels'.
For example suppose we have several plots to which we apply various amounts of Nitrogen fertiliser and then record the yield of maize. Let us suppose that we are going to examine the effect of applying Nitrogen at the rate of : 10kg, 20kg, 30kg, 40kg and 50kg.
| N=10, Y=20.5 | N=30, Y=43.1 | N=50, Y=46.4 |
| N=40, Y=45.0 | N=10, Y=21.6 | N=20, Y=36.4 |
| N=50, Y=48.4 | N=10, Y=22.9 | N=30, Y=44.4 |
| N=20, Y=38.5 | N=50, Y=47.3 | N=30, Y=44.2 |
| N=20, Y=38.0 | N=40, Y=45.5 | N=40, Y=46.1 |
We could consider Nitrogen to be a quantitative variable, to be fitted as a Regression covariate(s), or as a Classification factor with 5 'levels' or 'classes'. Run both these analyses and compute, and plot, the fitted values for each.
Possible models :
We could analyse this data as a one-way Analysis of Variance from a Completely Randomised Design. There would be the factor 'Nitrogen' with 5 levels. A suitable model would be :
Yij = µ + Ni + eij
where i = 1 to 5 levels of N
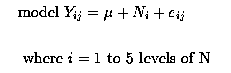
The SAS code would therefore be :
data crdreg1; input n wt; cards; 10 20.5 10 21.6 10 22.9 20 36.4 20 38.5 20 38.0 30 43.1 30 44.4 30 44.2 40 45.0 40 45.5 40 46.1 50 46.4 50 47.3 50 48.4 ; proc glm data=crdreg1; classes n; model wt = n; lsmeans n/stderr pdiff; run;
Compute the estimate of the mean for each Nitrogen level (the Least Squares Mean, i.e. µ + Nitrogen1, µ + Nitrogen2, etc).
SAS estimate statements estimate 'n = 10' intercept 1 n 1 0 0 0 0; estimate 'n = 20' intercept 1 n 0 1 0 0 0; etc..
We could also use the concept of contrasts to partition the Sums of Squares for the treatment (Nitrogen) into Linear, Quadratic, Cubic and Quartic components (see Steel, Torrie and Dickey, Ch15.7). This approach is reasonable and appropriate IF AND ONLY IF (IFF) the 'spacing' between the treatments (or the increment from one treatment level to the next) is the same. In this case, with 3 experimental units for all treatments we have a balanced design and the increment from one treatment (level) of Nitrogen to the next is the same (increments of 10kg N). The SAS contrasts are:
contrast 'Cubic and Quartic' n -1 2 0 -2 1,
n 1 -4 6 -4 1;
We could analyse this data as an Analysis of Covariance or Regression experiment with Nitrogen as regressions. Nitrogen could be fitted as linear, quadratic, cubic and quartic regressions. Thus a suitable model would be :
Yjk = µ + blkj + b1 Nk + b2 Nk2 + b3 Nk3 + b4 Nk4 + ejk
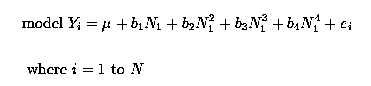
The SAS code would therefore be :
data crdreg1; input n wt; cards; 10 20.5 10 21.6 10 22.9 20 36.4 20 38.5 20 38.0 30 43.1 30 44.4 30 44.2 40 45.0 40 45.5 40 46.1 50 36.4 50 37.3 50 38.4 ; proc glm data=crdreg1; model wt = n n*n n*n*n n*n*n*n/solution; estimate 'n, linear ' n 1; estimate 'n, quadratic ' n*n 1; estimate 'n, cubic ' n*n*n 1; estimate 'n, quartic ' n*n*n*n 1; run;
Compute the estimate of the mean for each amount of Nitrogen (10 kg Nitrogen, 20 kg Nitrogen, etc). You should find that you have estimated exactly the same fitted value as previously when there were 5 classes.
SAS estimate statements estimate 'N=10' intercept 1 n 10 n*n 100 n*n*n 1000 n*n*n*n 10000; estimate 'N=20' intercept 1 n 20 n*n 400 n*n*n 8000 n*n*n*n 160000; etc..
We can compare the two models. We find that the Sums of Squares for the model are the same in both cases. If we compute the Contrast Sums of Squares for the joint test of all the regression parameters (4), with 4 d.f. we find that they are the same as the Sums of Squares for Nitrogen from the CRD design one-way ANOVA where Nitrogen had 5 levels, and hence 4 d.f. Think about this in the context of a graph where we have 2 points; with 2 points we need a straight line to provide a perfect fit to the the two points, i.e. 1 degree of freedom. If we had 3 points we know that a quadratic (linear+quadratic evidently!) will provide a perfect fit through three points, i.e. 2 degrees of freedom. With 4 points (levels) we will have 3 d.f., and a 3rd order polynomial (linear + quadratic + cubic) will be exactly equivalent. Likewise, with 5 classes (4 d.f.) a 4th order polynomial (linear + quadratic + cubic + quartic) is equivalent.
proc glm data=crdreg1;
model wt = n n*n n*n*n n*n*n*n;
contrast 'Regressions' n 1,
n*n 1,
n*n*n 1,
n*n*n*n 1;
We have 4 d.f., whether we have Nitrogen as 5 classes, with 4 d.f., or Nitrogen as a continuous quantitative trait with linear, quadratic, cubic and quartic regressions. The Sums of Squares for 'Regressions' (above) are equal to the Sums of Squares for Nitrogen (as 5 classes). So, fit a contrast for the Marginal Sums of Squares for the Cubic and Quartic regressions over and above the Linear and Quadratic :
contrast 'N3 and N4' n3 1,
n4 1;
These Sums of Squares are the Marginal Sums of Squares for adding both Cubic and Quartic regressions, over and above Linear and Quadratic. Now analyse this same data using a Regression model, but rather than fitting Linear, Quadratic, Cubic and Quartic regressions, fit only Linear and Quadratic. Compare the Residual Sums of Squares and d.f.e.. Also plot the fitted values for both cases (5 classes, and Linear and Quadratic regressions, graph.
Compute the difference between the Residual Sums of Squares for the model with only n and n*n, and the Residual Sums of Squares for the model with Nitrogen (as 5 classes). Do the same for the Residual degrees of freedom. What do you notice about these Sums of Squares and degrees of freedom?
Yes! They are the same as those for the joint contrast of the Cubic and Quartic regressions! Thus we can use this technique of fitting the same quantitative trait as classes or regressions to compare whether a simple Linear regression (or Linear + Quadratic regressions) provides a suitable fit vs. whether (5) classes provides a statistically significantly better fit. Linear regression (or Linear + Quadratic regressions) is a more parsimonious model (i.e. simpler), but depending upon the fitted values the class approach may, or may not, yield an improvement.
Analyses of Variance
| Source | d.f. | Sums of Squares | Mean Squares | F-ratio | Pr |
|---|---|---|---|---|---|
| SSRm | 4 | 1316.297 | 329.07 | 370.30 | 0.0001 |
| Residual | 10 | 8.887 | 0.889 |
| Source | d.f. | Sums of Squares | Mean Squares | F-ratio | Pr |
|---|---|---|---|---|---|
| SSRm | 4 | 1316.297 | 329.07 | 370.30 | 0.0001 |
| N, Type I | 1 | 1054.947 | 1054.947 | 1187.11 | 0.0001 |
| N2, Type I | 1 | 231.945 | 231.945 | 261.00 | 0.0001 |
| N3, Type I | 1 | 29.403 | 29.403 | 33.09 | 0.0002 |
| N4, Type I | 1 | 0.002 | 0.002 | 0.00 | 0.96 |
| Residual | 10 | 8.887 | 0.889 |
| Source | d.f. | Sums of Squares | Mean Squares |
|---|---|---|---|
| Regressions | 4 | 1316.297 | 329.07 |
| N3 and N4 | 2 | 29.405 | 14.703 |
| Source | d.f. | Sums of Squares | Mean Squares |
|---|---|---|---|
| SSRm | 2 | 1286.892 | |
| N, Type I | 1 | 1054.947 | 1054.947 |
| N2, Type I | 1 | 231.945 | 231.945 |
| Residual | 12 | 38.292 | 3.191 |
| Source | d.f. | Sums of Squares |
|---|---|---|
| Model 3 dfe | 12 | 38.292 |
| Model 1 dfe | 10 | 8.887 |
| 3 - 1 | 2 | 29.405 |
Thus we have obtained the degrees of freedom and Sums of Squares for the marginal improvement in going from a model with linear and quadratic regressions to a model with 5 classes (4 d.f.); using either Model 2, or Model 1 and Model 3 and computing the difference in d.f.e. and S.S.E. We can see that there is a statistically significant effect of the combined effect of the cubic and quartic regressions, or equivalently, that there is still a statistically significant improvement in going from a model with only linear and quadratic regressions to one with 5 classes (and hence 4 d.f.). Thus we conclude that Model 1 is the most appropriate. If the marginal effect of the cubic and quartic regressions had not been significant we would have concluded that there was no improvement in going from linear and quadratic regressions to 5 classes and that hence it would have been simpler, and more parsimonious, to use a model with linear and quadratic regressions.
These concepts can be extended to see whether simply a linear regression effect would have been adequate.
The approach detailed above is the classical orthogonal polynomial method detailed in Steel, Torrie and Dickey, and inumerable other statistical textbooks. However, as mentioned above, it has the limitation that if the intervals are not equal then it becomes difficult to compute what the coefficients should be. In addition, if we have a factor with several/many levels then the order of the polynomial can rapidly get out of hand, and in addition it will be very easy for us to accidently miss out one of the regressions. A technique that is easily used is to exploit the fact that SAS PROC GLM (and PROC MIXED) allow us to 'overfit' a model, or over-parameterise, by fitting both regressions and classes of the same effect. Obviously we cannot simply fit the variable n as both a classification variable and a regression covariate, because if we specify in PROC GLM that n is a class variable then it cannot simultaneously be a regression covariate. What we can do is create a second variable with exactly the same value as n and then use that as a classification variable and n as the regression. Thus, suppose that we write the model as:
Where Ni is the linear regression covariate and b1 is the linear regression coefficient; and Ni2 is the quadratic regression covariate and b2 is the quadratic regression coefficient. grpi is the effect of group (Nitrogen treated as 5 classes, 4 d.f.) over and above the effect of the Linear and Quadratic regression effects.
We have to create the second variable in the SAS data step, thus the SAS code would therefore be :
data crdreg1; input n wt; grp = n; cards; 10 20.5 10 21.6 10 22.9 20 36.4 20 38.5 20 38.0 30 43.1 30 44.4 30 44.2 40 45.0 40 45.5 40 46.1 50 36.4 50 37.3 50 38.4 ; proc glm data=crdreg1; classes grp; model wt = n n*n grp; run;
Here is another data set to analyse to compare and contrast with the first example. Repeat the analyses, calculations, estimation of fitted values and plotting of points. A group of 15 dairy cows were fed various amounts of feed and their milk yield recorded:
Data Independent Dependent
variable variable
Cow Kg feed Amount of milk
1 20 11.1
2 20 16.6
3 20 15.6
4 30 18.1
5 30 22.8
6 30 21.6
7 40 25.2
8 40 30.1
9 40 26.7
10 50 28.6
11 50 33.7
12 50 35.8
13 60 34.5
14 60 31.4
15 60 33.6
After comparing analyses from this data what are your conclusions?

Additional references
Steel, Torrie and Dickey. Ch. 15.6 and 15.7