See Steel, Torrie and Dickey, Chapter 14. Also review Chapters 10, 11, 12 and 13.
This first problem in multiple regression is taken from Steel, Torrie and Dickey, Chapter 14, Table 14.1. Nitrogen %, Chlorine % and Potassium % were measured in soil samples taken from various plots of tobacco plants. For each plot the leaf burn was measured. The natural logarithm of the leaf burn was used as the dependent variable. The object of the study was to see if the above mentioned factors (N%, Cl% and K%) affected leaf burn.
Alternatively, if one wishes to consider an example using animals let us imagine the same basic data, but supposing that we had 30 cows and we were conducting an experiment to look at the effects of Energy density of the diet and Acid Base Balance (Chlorine % and Potassium %) on milk yield. The cows are randomly assigned and fed different diets each with various Energy densities and differing amounts of Chlorine % and Potassium % and the milk yield for each cow is recorded.
Note that this data is the same as that described earlier in the 'Introduction to the SAS Language'.
| Nitrogen % | Chlorine % | Potassium % | Log leaf burn |
|---|---|---|---|
| Energy Density | Chlorine % | Potassium % | Milk Yield |
| 3.05 | 1.45 | 5.67 | 0.34 |
| 4.22 | 1.35 | 4.86 | 0.11 |
| 3.34 | 0.26 | 4.19 | 0.38 |
| 3.77 | 0.23 | 4.42 | 0.68 |
| 3.52 | 1.10 | 3.17 | 0.18 |
| 3.54 | 0.76 | 2.76 | 0.0 |
| 3.74 | 1.59 | 3.81 | 0.08 |
| 3.78 | 0.39 | 3.23 | 0.11 |
| 2.92 | 0.39 | 5.44 | 1.53 |
| 3.10 | 0.64 | 6.16 | 0.77 |
| 2.86 | 0.82 | 5.48 | 1.17 |
| 2.78 | 0.64 | 4.62 | 1.01 |
| 2.22 | 0.85 | 4.49 | 0.89 |
| 2.67 | 0.90 | 5.59 | 1.40 |
| 3.12 | 0.92 | 5.86 | 1.05 |
| 3.03 | 0.97 | 6.60 | 1.15 |
| 2.45 | 0.18 | 4.51 | 1.49 |
| 4.12 | 0.62 | 5.31 | 0.51 |
| 4.61 | 0.51 | 5.16 | 0.18 |
| 3.94 | 0.45 | 4.45 | 0.34 |
| 4.12 | 1.79 | 6.17 | 0.36 |
| 2.93 | 0.25 | 3.38 | 0.89 |
| 2.66 | 0.31 | 3.51 | 0.91 |
| 3.17 | 0.20 | 3.08 | 0.92 |
| 2.79 | 0.24 | 3.98 | 1.35 |
| 2.61 | 0.20 | 3.64 | 1.33 |
| 3.74 | 2.27 | 6.50 | 0.23 |
| 3.13 | 1.48 | 4.28 | 0.26 |
| 3.49 | 0.25 | 4.71 | 0.73 |
| 2.94 | 2.22 | 4.58 | 0.23 |
Thus a suitable linear model involving the 3 regression effects would be :
model Yi = b0 + b1 Xi1 + b2 Xi2 + b3 Xi3 + animali + ei
![]()
We have only 1 measurement/animal. Therefore we cannot seperate the effect animali (the random effect of animal i) from ei (the random error associated with measurement i). Thus the effects animali and ei are confounded one with another and will be lumped together as one combined term, ei, where ei = animali + random errori. Thus we will re-write the model as
model Yi = b0 + b1 Xi1 + b2 Xi2 + b3 Xi3 + animali + ei
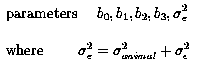
SAS code, data step, PROC REG, PROC GLM and PROC IML.
| Source of variation | d.f. | Sums of Squares | Mean Squares | F-ratio | Pr |
|---|---|---|---|---|---|
| Total | 30 | 20.8074 | |||
| Model, SSR | 4 | 19.6226 | |||
| C.F. for mean | 1 | 14.1179 | |||
| Model after µ | 3 | 5.5047 | 1.8349 | 40.27 | 0.0001 |
| R(b1| µ, b2, b3) | 1 | 2.6587 | 2.6587 | 58.35 | 0.0001 |
| R(b2| µ, b1, b3) | 1 | 1.6511 | 1.6511 | 36.23 | 0.0001 |
| R(b3| µ, b1, b2) | 1 | 1.2049 | 1.2049 | 26.44 | 0.0001 |
| Residual | 26 | 1.1848 | 0.0456 |
If we want to see in more detail the actual output from SAS (formatted as HTML Web pages) we can look at:
X'X matrix, augmented with X'Y and Y'Y
(X'X)-1 matrix, augmented with the estimates for b, and SSE
These are produced by the XPX and INVERSE (I) options on the model statement of PROC GLM.
proc glm data=reg1; model y = x1 x2 x3/xpx i solution; run;
The above code runs the procedure glm (General Linear Model) using the SAS dataset reg1. The model statement tells us that the dependent variable is the SAS variable y (which must obviously exist in our data set reg1) and the the explanatory, independent variables are x1, x2 and x2. We also want the options (specified by the /....) XPX for the X`X matrix, I for the (X`X)-1 matrix and SOLUTION for the vector of estimates (solutions), standard errors of the estimates, tcalc (calculated t-value), and the probability of obtaining such a large t-value just by chance when the Null Hypothesis is true.
Note the SAS output ANOVA table corresponds in part to that given above, except that what SAS, in common with many other statistical packages, calls the Model is in fact the Model after corrrecting for the Mean. The Type III Sums of Squares correspond to those given above, they are the the Marginal Sums of Squares for each effect, that is to say they are the Sums of Squares for X1 adjusted for X2 and X3 (the other terms in the model), the Sums of Squares for X2 adjusted for X1 and X3, etc.
When we ran the PROC GLM model we actually had more statements that simply the 3 lines shown above; we also made use of the OUTPUT statement of GLM.
proc glm data=reg1; /* Using PROC GLM (General Linear Model) */
/* note xpx, i and solution provide us with
X transpose X, its inverse and the solutions */
model y = x1 x2 x3/xpx i solution;
/* compute and output fitted/predicted values (p=yhat), residuals (r=ehat),
standard errors of the predicted values (stdp=se) and 95% upper and
lower confidence limits of our fitted values (u95m=ul and l95m=ll) */
output out=reg1p p=yhat r=ehat stdp=se u95m=ul l95m=ll;
run;
quit; /* Exit PROC GLM */
Here we specify that we want to "output" to a new SAS data set some results from the GLM procedure (out=reg1p), where reg1p is the name of the NEW data set to be created. We want to store the predicted (fitted, or estimated) values in a new variable yhat (p=yhat) and the estimated residuals in a new variable ehat (r=ehat). The p= and r= SAS keywords are required, the names that I have given to the Predicted, fitted values (yhat) and Residuals (ehat) are my arbitrary choice; however, it just seemed to make sense to me to choose names that are close to what they represent. If I had chosen to write p=fred that would have been quite acceptable to SAS, but later if I looked at the data set reg1p I might have absolutely no idea what the variable fred represents!!!
Then the following SAS statements invoke (run) the print procedure to 'print' output the newly created SAS data set reg1p, which contains the dependent variable (y), the explanatory variables from out model (x1, x2 and x3), as well as the fitted values (y hat), the residuals (e hat) as well as the standard error of each of our estimated (fitted/predicted) values, and the estimates of the upper and lower 95% confidence interval. SAS output from PROC PRINT.
/* print out the new data set reg1p */ proc print data=reg1p; title 'Observations, fitted values, residuals etc'; run;
We can plot the results from this analysis using the high quality graphics procedures of SAS/GRAPH;
proc gplot data=reg1p; title ' Graph of y vs X1 '; plot y*X1; run; proc gplot data=reg1p; title ' Graph residuals vs. X1 '; plot ehat*X1; run;
Note: an important point here that will come back again and again and that we can, and should, exploit is the fact that it is ALWAYS possible to fit different models and compare them via their effects on the residual Sums of Squares and degrees of freedom. For example, if we fit a model as above with X1, X2 and X3 as parameters in the model we shall obtain a certain Sums of Squares and degrees of freedom for the Residual. Let us call this the 'Full Model'. Then suppose that we were to fit a model without X1 in it. This would be equivalent to saying that the parameter X1 (or at least the regression coefficient b1 associated with it) is equal to Zero; i.e. has not effect. Let us call this the 'Reduced Model'. Then the effect of X1 can be computed as the change in the residual Sums of Squares and degrees of freedom.
| d.f.e | Sums of Squares Error | |
|---|---|---|
| Reduced Model | 27 | 3.8435 |
| Full Model | 26 | 1.1848 |
| Difference, R(X1 | b0, X2, X3) | 1 | 2.6587 |
Note that this is exactly the same as the marginal Sums of Squares that we computed for X1 using SAS/GLM. This is not an accidental 'fluke'; it's the real thing. If we want to we can always to this; fit a model with a parameter and fit another model without the parameter (equivalent to setting it to Zero), what is known as a reduced model, and then obtain the degrees of freedom and Sums of Squares from differencing the d.f.e and SSE's.
